Hi Benching Community, The Molecular Biology team is very excited to introduce our new combinatorial cloning tool!
Crucial to our customers’ sequence design workflows, cloning is often and increasingly conducted at scale — both in silico and at the bench. Knowing that, we set out to design a tool that would enable scientists to seamlessly and reliably model combinatorial assembly in Benchling, all within the same platform where you orchestrate and track the rest of your lab work.
With our new combinatorial cloning tool, you can:
- Design many constructs at once with an intuitive interface (and one that, we think, is actually fun to use!)
- Visualize fragments and constructs as you’re designing them
- Choose from multiple cloning methods including Golden Gate and Gibson
- Customize assemblies by limiting constructs to desired combinations of fragments and reusing primers
- Reference a record of your assembly to trace how constructs were designed
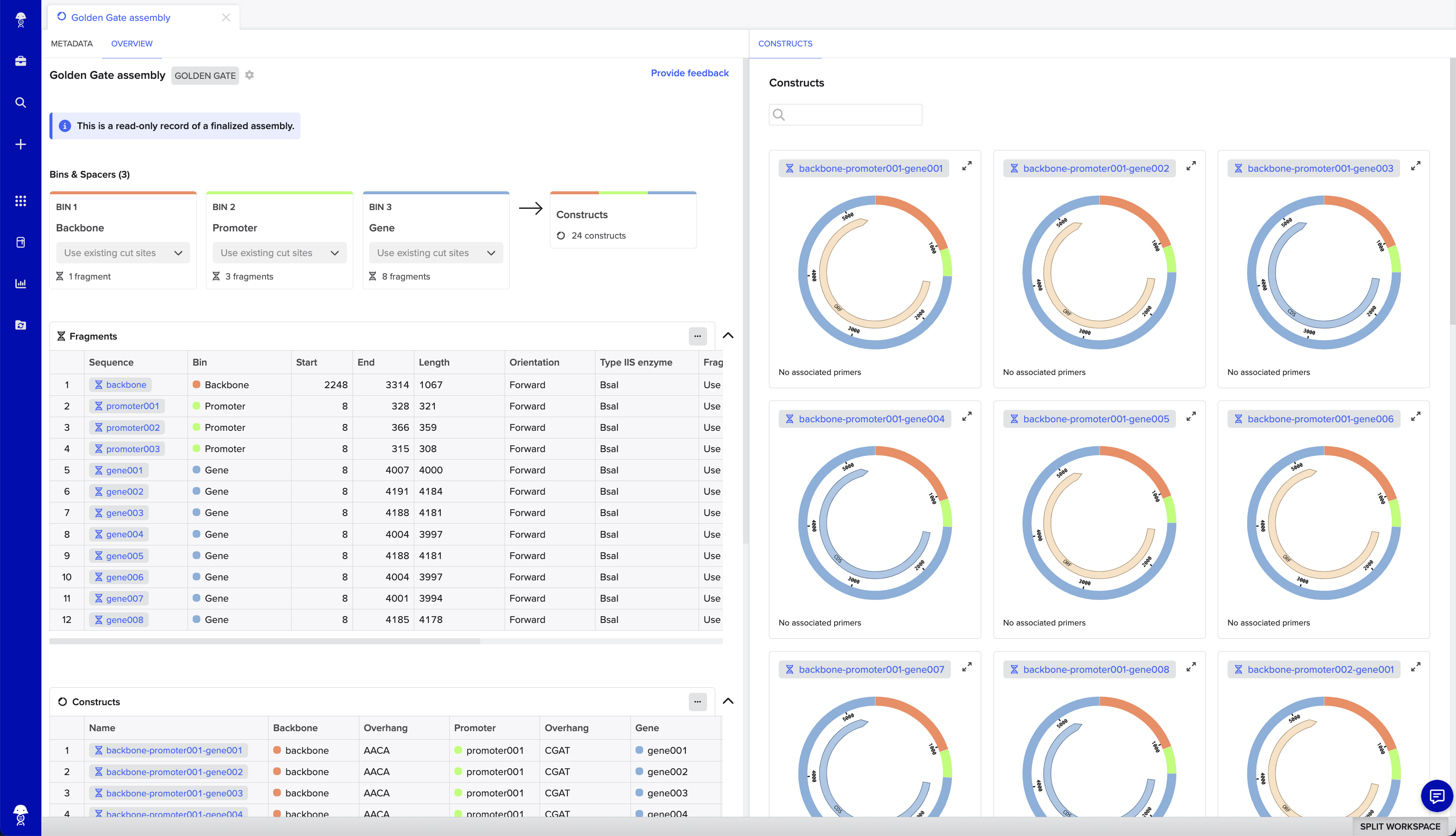
To access the tool, find + in the left-hand navigation menu > Assembly > Assemble DNA sequences by cloning and select your assembly settings.
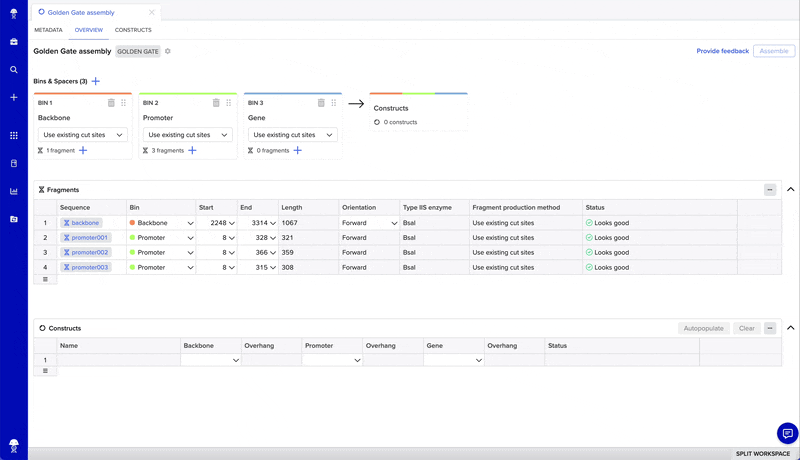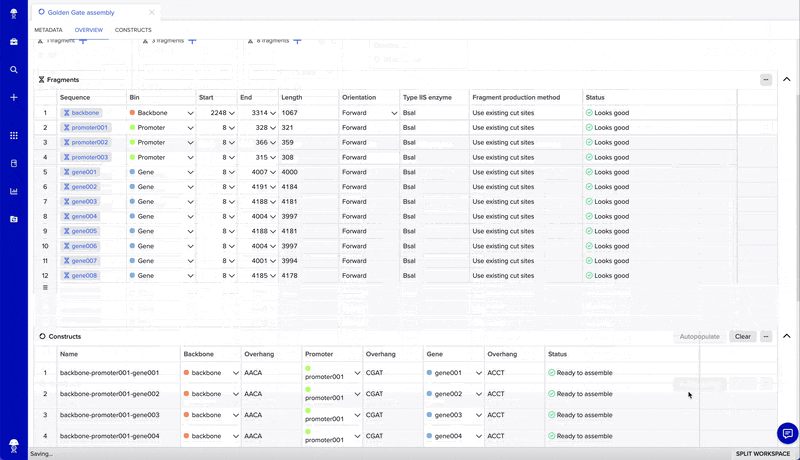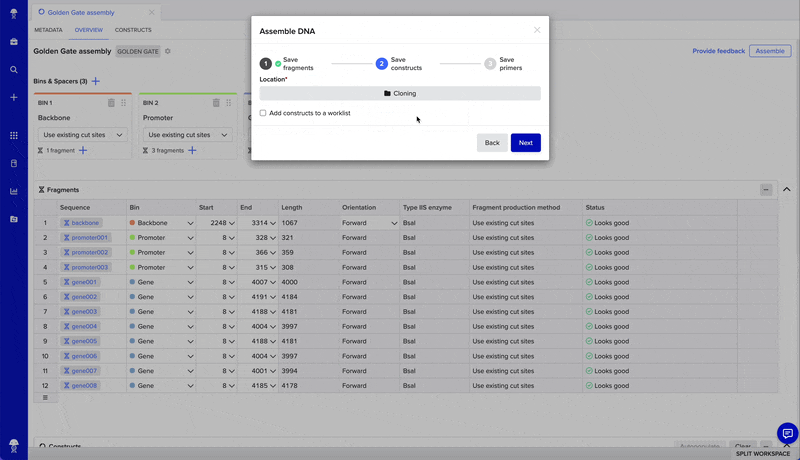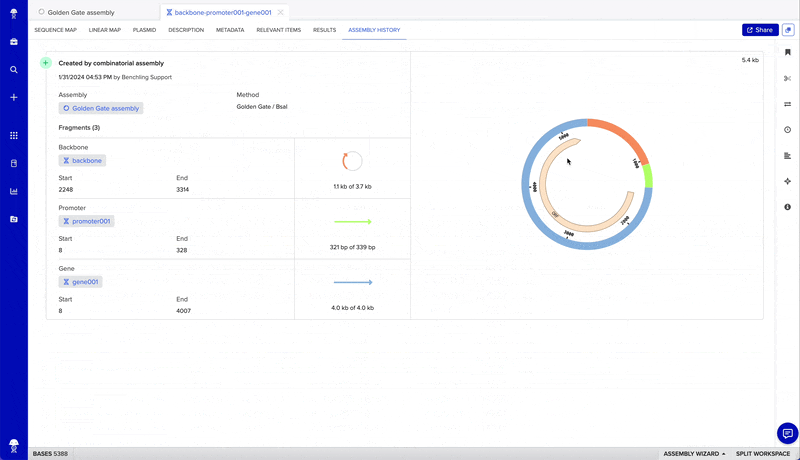
From there, you can design constructs in four steps:
- Add and adjust bins (one bin for every insert plus one for your backbone)
- Add fragments to bins using the "+" on each bin and the fragments table
- Preview constructs by selecting “autopopulate” or manually filling the constructs table
- Finalize your assembly to create new sequences
For more details, check out these step-by-step guides for Golden Gate, Gibson, and homology-based assembly in Benchling.
While this tool is designed to replace legacy bulk assembly, the legacy tool will be available for a limited time. We will notify you before it becomes unavailable.
As you transition to our new combinatorial cloning tool, we welcome any and all feedback. Our goal is to design molecular biology tools that are easy to use, save you time, and most importantly, match the reality and complexity of your science. Your feedback helps us ensure that we’ve been able to do that and influences how we continue to develop this tool over the coming months. Next, we are adding API support for assemblies (finally, we know!); assembly endpoints will be available later this spring.
And in case you missed it — last year, we released a combinatorial “concatenation” tool that uses the same patterns as our cloning tool. Combinatorial concatenation allows you modularly design DNA, RNA, or AA by joining sequences end to end. This tool can also be accessed from the Assembly sub-menu (Assembly > Assemble sequences and oligos by concatenation).
Happy assembling!